Good news: CEGMA is more popular than ever — Bad news: CEGMA is more popular than ever
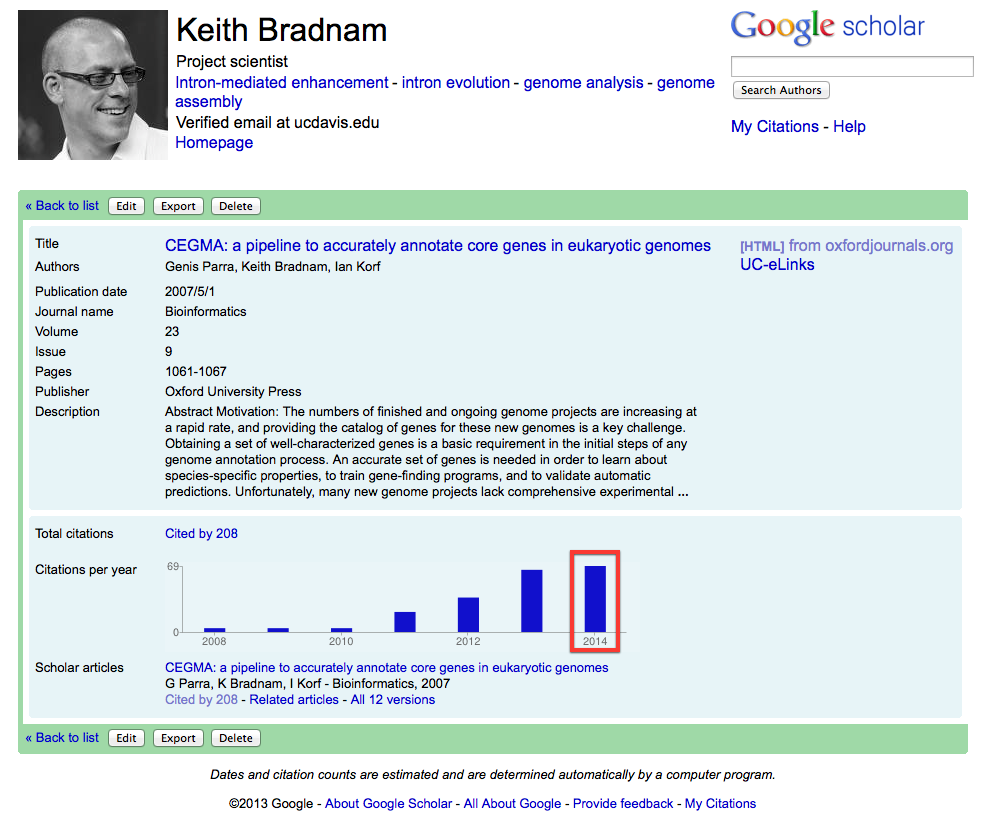
I noticed from my Google Scholar page today that our 2007 CEGMA paper continues to gain more and more citations. It turns out that there have now been more citations to this paper in 2014 than in any previous year (69 so far and we still have almost half a year to go):
I've previously written about the problems of supporting software that a) was written by someone else and b) is based on an underlying dataset that is now over a decade old. These problems are not getting any easier to deal with.
In a typical week I receive 3–5 emails relating to CEGMA; these are mostly requests for help with installing and/or running CEGMA, but we also receive bug reports and feature requests. We hope to shortly announce something that will help with the most common problem, that of getting CEGMA to work. We are putting together a virtual machine that will come pre-installed and configured to run CEGMA. So you'll just need to install something like VirtualBox, and then download the CEGMA VM. Hopefully we can make this available in the coming week or two.
Unfortunately, we have almost zero resources to devote to the continuing development of this old version of CEGMA; any development that does happen is therefore extremely limited (and slow). A forthcoming grant submission will request resources to completely redevelop CEGMA and add many new capabilities. If this grant is not successful then we may need to consider holding some sort of memorial service for CEGMA as it becoming untenable to support the old code base. Seven years of usage in bioinformatics is a pretty good run and the website link in the original paper still works (how many other bioinformatics papers can claim this I wonder?).
Update: 2014-07-21 14.44
Shaun Jackman (@sjackman on twitter) helpfully reminded me that CEGMA is available as a homebrew package. There is also an iPlant application for CEGMA. I've added details of both of these to a new item in the CEGMA FAQ:
Update: 2014-07-22 07.36
Since publishing this post, I've been contacted by three different people who have pointed out different ways to get CEGMA running. I'm really glad that I blogged about this else I may not have found about these other methods.
In addition to Shaun's suggestion (above), it seems that you can also install CEGMA on Linux using the Local Package Manager software. Thanks to Masahiro Kasahara for bringing this to my attention. Finally, Matt MacManes alerted me to the fact that their is a public Amazon Machine Instance called CEGMA on the Cloud. More details here.
Update: 2014-07-30 19.31
Thanks to Rob Syme, there is now a Docker container for CEGMA. And finally, we have now made a Ubuntu VM that is pre-installed with CEGMA (thanks to Richard Feltstykket at the UC Davis Genome Center's Bioinformatics Core).